Proposal 7: ARTA

Antimicrobial Resistance: routes of Transport in the gulf of Amvrakikos
When: July 2022
Where: Gulf of Amvrakikos
What: Seawater sampling for collecting microbial DNA
Who: National Institute of Oceanography and Applied Geophysics – OGS, Trieste, Italy & Hellenic Center for Marine Research (HCMR) Heraklion and Anavyssos, Greece
In the ARTA research cruise (funded by "RePhil"), the R/V PHILIA was utilized to collect seawater and riverine water samples in order to detect the routes of transport of antibiotic resistant bacteria.
In fact, due to the massive utilization of antibiotics, the development of bacteria that have acquired specific genes to defend themselves from drugs is a phenomenon of widespread concern. Since one of the sources of antibiotic resistant bacteria in coastal areas is represented by freshwater inputs, the Amvrakikos Gulf was chosen as a study area, being affected by the outflows of the rivers Arachthos and Louros.
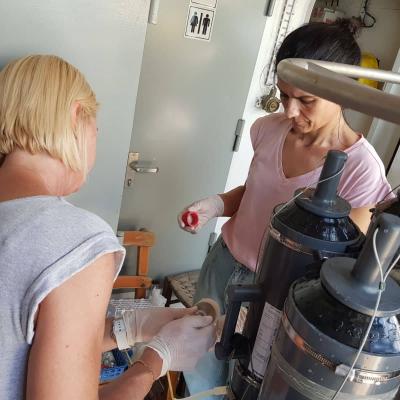
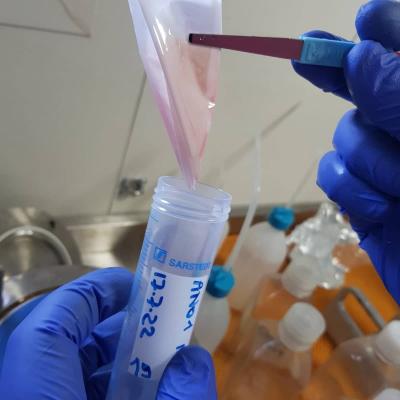
The activities onboard consisted of the characterization of the physical structure of the water column through CTD casts (deployment of a multiparametric probe for the real-time acquisition of temperature, salinity, chlorophyll a fluorescence and dissolved oxygen concentration) at increasing distances from the mouth of the aforementioned rivers. At the same stations, seawater was sampled at different depth layers for the collection of bacterial DNA (potentially bearing bacterial antibiotic-resistance genes).
Furthermore, water samples for biogeochemical (nutrients, particulate matter, etc.) and biological (quali-quantitative determination of bacterial, eukaryotic and viral assemblages) analyses were collected to provide a robust background to the molecular analyses.
The availability of a tender also allowed the scientific party to sample in the shallow river mouth and upstream areas.
In the ARTA research cruise (funded by "RePhil"), the R/V PHILIA was utilized to collect seawater and riverine water samples in order to detect the routes of transport of antibiotic resistant bacteria.
In fact, due to the massive utilization of antibiotics, the development of bacteria that have acquired specific genes to defend themselves from drugs is a phenomenon of widespread concern. Since one of the sources of antibiotic resistant bacteria in coastal areas is represented by freshwater inputs, the Amvrakikos Gulf was chosen as a study area, being affected by the outflows of the rivers Arachthos and Louros.
The activities onboard consisted of the characterization of the physical structure of the water column through CTD casts (deployment of a multiparametric probe for the real-time acquisition of temperature, salinity, chlorophyll a fluorescence and dissolved oxygen concentration) at increasing distances from the mouth of the aforementioned rivers. At the same stations, seawater was sampled at different depth layers for the collection of bacterial DNA (potentially bearing bacterial antibiotic-resistance genes).
Furthermore, water samples for biogeochemical (nutrients, particulate matter, etc.) and biological (quali-quantitative determination of bacterial, eukaryotic and viral assemblages) analyses were collected to provide a robust background to the molecular analyses.
The availability of a tender also allowed the scientific party to sample in the shallow river mouth and upstream areas.